The DNA molecule is the genetic material and can be visualised as a polymer of a nucleotide monomer.
DNA – A BRIEF HISTORY
Friedrich Miescher, in 1868 was the first person to isolate and characterise DNA. He named this Phosphorous-containing substance “Nuclein“.
Later, Oswald T Avery, Maclyn McCarty, and Colin Macleod conducted studies using virulent and non-virulent strains of Streptococcus pneumonia and concluded that the DNA was the genetic material.
Alfred D. Hershey and Martha Chase removed the remaining doubts regarding DNA in 1952 and provided evidence to prove that DNA is the genetic material.
The experiment of Erwin Chargaff and his colleagues led him to construct the following conclusions.
- The DNA’s base composition generally varies from one species to another species.
- DNA specimens separated from different tissues of the same species have the same base composition.
- The base composition in a given species does not change with an organism’s age, or any other factors will not alter the base composition.
- Regardless of the species, the number of adenosine residues equals the number of thymidine residues (A=T). The number of guanosine residues equals the number of cytidine residues (G=C). In short, A+G=T+C.
These relationships are known as Chargaff’s rules.
Rosalind Franklin and Maurice Wilkins of Kings College used the X-ray diffraction method to produce a DNA diffraction pattern. This pattern concluded that DNA molecules are helical with two periodicities along their axis, a primary one of 3.4 amstrong and a secondary of 34 amstrong.
DNA STRUCTURE OR DOUBLE HELIX MODEL
In 1953, James Watson and Francis Crick used the data from X-ray diffraction to postulate a three-dimensional model of DNA structure.
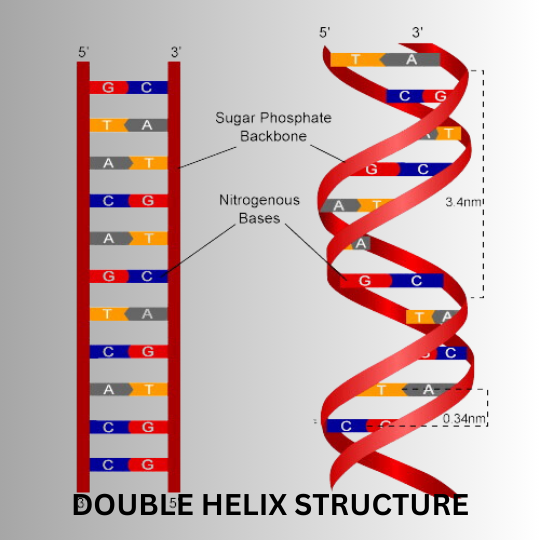
The proposed structure of DNA included the following elements.
- The DNA molecule is composed of two chains of nucleotides. This conclusion is based on Linus Pauling’s proposal, but Pauling had presented that DNA was composed of three nucleotide strands.
- The two chains twist around each other to form a pair of right-handed helices, and when the observer looks down the central axis, the molecule would see that each strand follows a clockwise path as it moves away from the observer. The spots produced by the Rosalind Franklin x-ray pattern revealed the helical nature.
- The two chains of DNA are anti-parallel. If one chain is aligned in a 3′ to 5′ direction, then the other chain should be aligned in a 5′ to 3′ direction.
- The sugar-phosphate backbone of each strand is located on the exterior of the DNA molecule, with two sets of bases projecting toward the centre.
- The bases of the DNA molecule are stacked or arranged by Hydrophobic interaction and Van de Waals forces.
- Hydrogen bonds maintain the two strands of DNA molecule together. Even though the individual hydrogen bonds are weak, the strength of hydrogen bonds is addictive, and a large number of hydrogen bonds together will make the molecule structure stable.
- The distance from the backbone’s phosphorus atom to the axis’s centre is 1 nm. So, the width of the double helix is 2nm.
- A purine in one chain is always paired with a pyrimidine in the other chain because the nitrogen atom linked to carbon 4 of cytosine and carbon 6 of adenine are predominantly in the amino configuration(NH2). The oxygen atom linked to carbon 6 of guanine and carbon 4 of thymine is predominantly in a keto (C=O) configuration rather than the enol (C-OH) form. These structural restrictions on the configurations of the bases suggested that adenine was the only purine structurally capable of bonding to thymine and guanine with cytosine.
- A-T and G-C base pairs had the same geometry, so there were no restrictions on the sequence of bases.
- Around the outer surface, there will be a wider major groove and a more narrow minor groove.
- The DNA double helix molecule makes one complete turn every ten residues, i.e., 3.4 nm or 150 turns per million daltons of molecular mass.
- The two chains of DNA molecules will be complementary to one another.
THE UNUSUAL STRUCTURE OF DNA
B- DNA
The B form of DNA is the most stable structure under normal conditions. It is right-handed and has a diameter of 20 amstrong. B-form DNA has an anti-glycosyl bond conformation. The base pairs lie in a plane that is close to perpendicular to the helix axis.
A- DNA
A-DNA is created when the B-form DNA is dehydrated ( the relative humidity is reduced from 92% to 75%, and sodium, potassium, and Cs+ ions are present in the medium). The helix pitch (helix in DNA moves a total distance) is 25.3 amstrong. The base pairs have a marked propeller twist concerning the helix axis. It has an anti-glycosyl bond conformation.
Z- DNA
The helix is left-handed, with the phosphates in the DNA backbone in a zigzag manner. It was discovered by RICH, Nordheim, and Wang in 1984.
Here the diameter is 20 amstrong. It has an anti-glycosyl bond conformation for pyrimidine and syn for purines( Due to Steric constraints, purines in purine nucleotides are restricted to two stable conformations concerning deoxyribose, called syn and anti. Pyrimidines are generally limited to the anti-conformation because of steric interferences between the sugar and the carbonyl oxygen at C-2 of the pyrimidine).
D-DNA
C- DNA is formed at 66% relative humidity in the presence of Li+ ions. The diameter is smaller than that of both B-DNA and A-DNA. It is also a right-handed one.
C-DNA
It is extremely rare, with only eight base pairs per helical turn and devoid of guanine.
A, B, and C forms are found in all DNA molecules irrespective of their base sequence.
PALINDROMIC DNA
The region of DNA with inverted repeats of base sequence has two-fold symmetry over strands of DNA. They are self-complementary within each strand and, therefore, have the potential to form Hairpin or Cruciform structures.
BENT DNA
This type of DNA is created whenever four or more adenine residues occur sequentially in one of the two strands. Six adenosines in a row can produce a bend of about 18 degrees. This bending is vital in the binding of some proteins.
HOOGSTEEN POSITIONS
The atoms that participate in the hydrogen bonding of triplex DNA are often called Hoogsteen positions. The non-Watson-Crick pairing is known as the Hoogsteen pairing. Karst Hoogsteen first recognised this pairing in 1963. Hoogsteein pairing allows the formation of triplex DNAs, which are more stable in low pH.
H-DNA
It is usually found in polypyrimidine or polypurine segments that contain a mirror repeat within themselves. An essential feature of H-DNA is pairing and interwinding three strands of DNA to form a triplex helix.
SINGLE-STRANDED DNA
Robert Sinsheimer first identified it in 1959 in φX174, a small virus that infects E. Coli (It is single-stranded only for part of its life, and after infecting E. Coli, it will become double-stranded). Here, DNA is single-stranded instead of double-stranded. It behaves as a randomly coiled polymer.



